Linking spatial & temporal Scales in biology
The availability of completely sequenced genomes and genome-scale datasets provides us with a wealth of information and the opportunity to investigate fundamental problems of a different magnitude and kind that is complementary to conventional approaches. While much of this information is publicly available, one of the major challenges we face is to be able to mine this enormous amount of data and integrate them in a meaningful way in order to (i) generate experimentally testable hypotheses and (ii) uncover general principles that govern living systems.
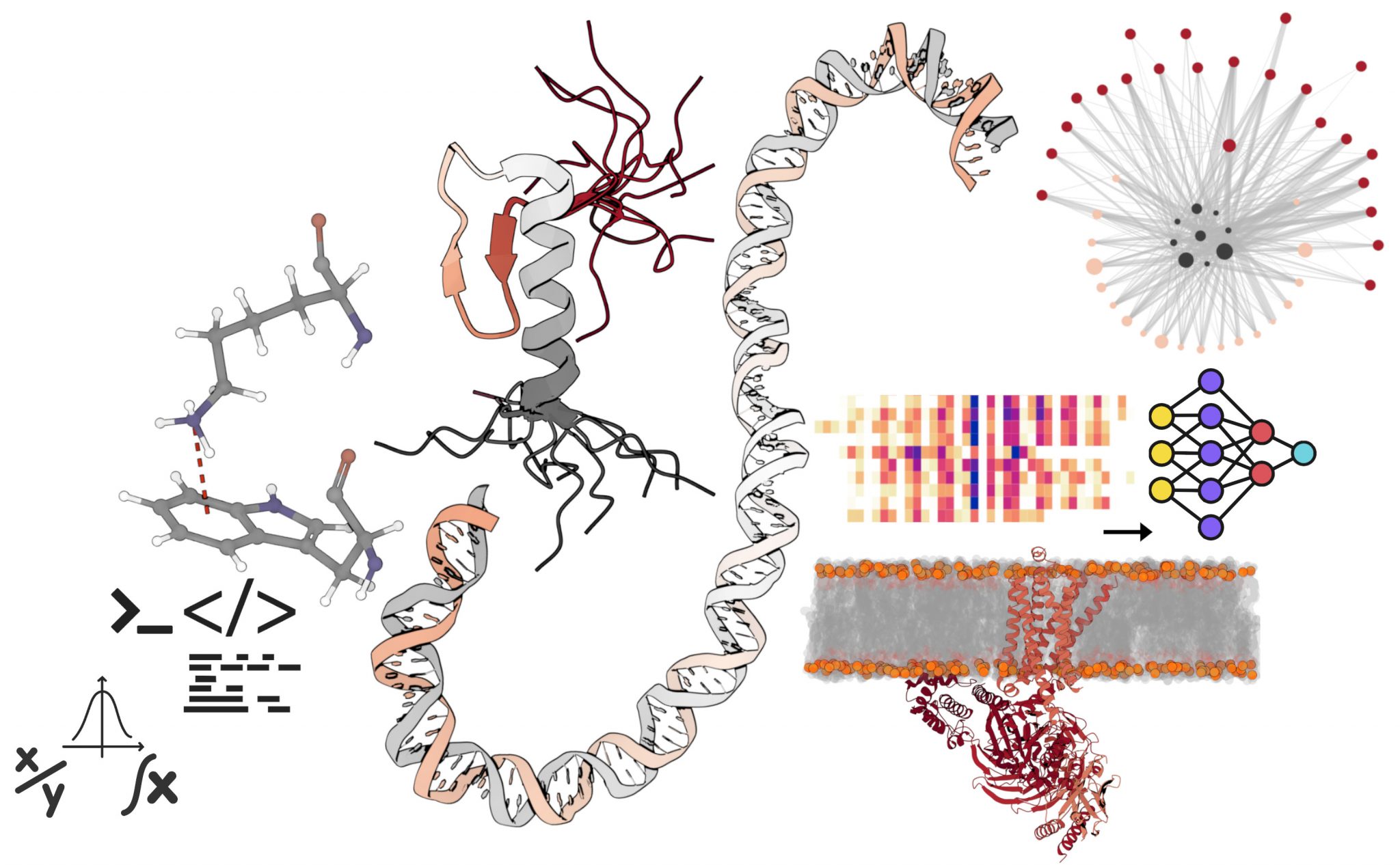
Research in my group is aimed at understanding how regulation is achieved at different scales of complexity (Molecular, Systems and Genome level) in cellular systems and how they influence genome evolution. This area of research is important because many human diseases are a result of dysfunction of regulatory processes and understanding design principles of regulation can be exploited in biotechnology and medicine.
Regulation at the molecular level
At the molecular level, we aim to investigate regulatory proteins that are involved in protein-protein, protein-nucleic acid and protein-small molecule interactions. In particular, we aim to gain an understanding of the proteins involved in (a) GPCR receptor signalling, (b) chromatin modification, and (c) ubiquitin pathways.
Regulation at the systems level
At the systems level, we aim to gain an understanding of (a) how aggregation prone proteins are tolerated and regulated, (b) how pathogens could possibly exploit host machinery and signalling by secreting unstructured proteins and (c) understand the regulation and dynamics of proteins within the context of their position in the various molecular interaction networks.
Regulation at the genome level
At the genome level, we investigate (a) the interplay between nucleosome modification and genome organization, (b) mutations and their impact on genome and transcriptome evolution, (c) the genetic basis of natural behavioural variation and (d) bacterial transcription networks and genome evolution.
To pursue these lines of research, we develop new computational methods that will make use of the publicly available large-scale datasets. These include sequence, structure, expression and molecular interaction data from several different model organisms ranging from bacteria to fungi to humans. We also exploit newly available data on natural variation, cancer genome sequence and gene expression and nucleosome modification data, and information from genetic screens linking genotype with phenotype. The problems that we work on in my group have the ultimate objective of being able to identify new regulatory principles and features of regulatory systems, to exploit our findings and the general principles that we describe in specific applications such as genetic engineering, and to better understand regulatory dysfunctions leading to human diseases.