Tilman Flock is awarded the Max Perutz Student Prize 2015
Big congratulations to Tilman to win this year’s Max Perutz Student Prize.
The Max Perutz Student Prize is awarded annually for outstanding work performed at the LMB prior to the award of a PhD. The 2015 prize has been awarded to Tilman Flock, for his comprehensive computational analysis of GTP-binding proteins (G proteins), revealing the universal nature of their interactions and activation.
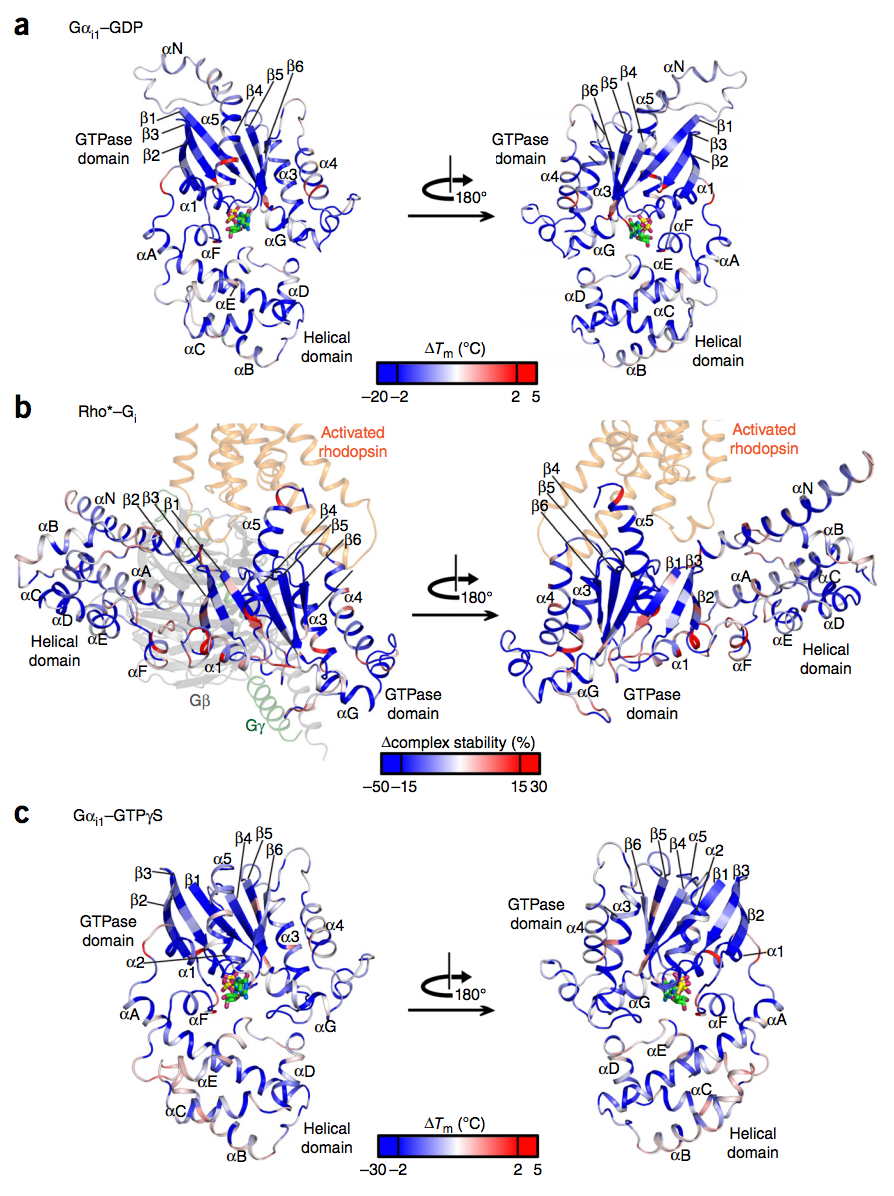
Tilman, a third year PhD student in the group of Madan Babu in the Structural Studies Division, undertook a systematic analysis of over 80 structures and 950 sequences of G proteins from different species to reveal how the core mechanism of activation and recognition is conserved, even while new specific interactions evolve. In humans there are over 800 G protein-coupled receptors which, upon binding of an extracellular ligand, activate one or more of 16 different G proteins by triggering the exchange of GDP for GTP, thus initiating a series of signalling pathways. It is these receptors that allow us to smell different chemicals, respond to adrenalin, and sense neurotransmitters in the brain, amongst many other functions. More than 30% of all prescribed small molecule drugs act by stimulating or inhibiting them. Since the 1980’s, there have been over 11,000 publications relating to how G proteins work. Tilman’s elegant methodological approach unifies a vast amount of data, providing a framework for the whole field. It is a general approach that can be extended to other proteins, and it shows the power of large-scale analysis of the ever-growing mass of published data.
See Flock et al, Nature 524, 173-179 (2015).
The research student prize is awarded by the Max Perutz Fund. The Fund was established for the promotion and advancement of education and research in molecular biology and allied biomedical sciences.

Photo and text are taken from the LMB website.