admin
May 5, 2016
5:03 pm
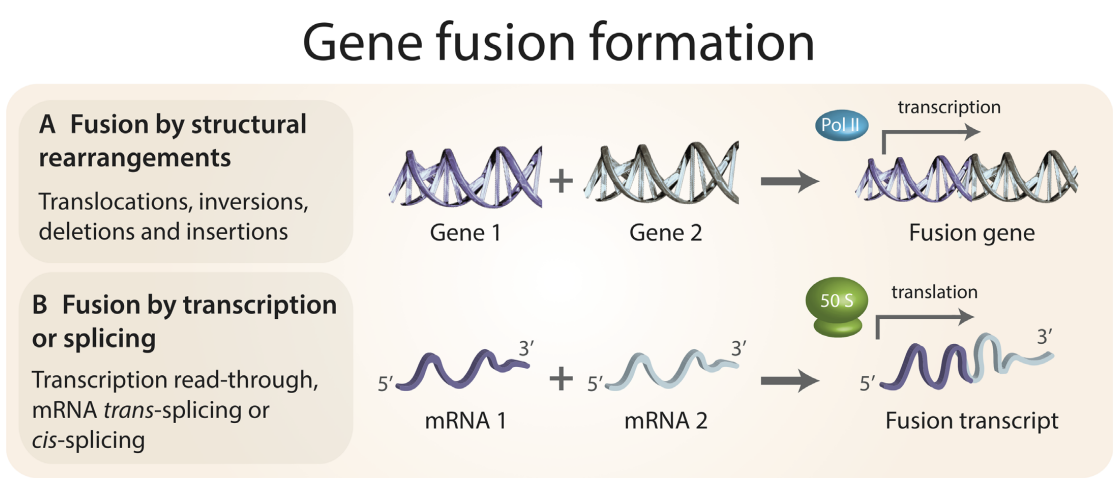
Although gene fusions have been recognized as important drivers of cancer for decades, our understanding of the prevalence and function of gene fusions has been revolutionized by the rise of next-generation sequencing, advances in bioinformatics theory and an increasing capacity for large-scale computational biology. The computational work on gene fusions has been vastly diverse, and the present state of the literature is fragmented. It will be fruitful to merge three camps of gene fusion bioinformatics that appear to rarely cross over: (i) data-intensive computational work characterizing the molecular biology of gene fusions; (ii) development research on fusion detection tools, candidate fusion prioritization algorithms and dedicated fusion databases and (iii) clinical research that seeks to either therapeutically target fusion transcripts and proteins or leverages advances in detection tools to perform large-scale surveys of gene fusion landscapes in specific cancer types. In this review, we unify these different-yet highly complementary and symbiotic-approaches with the view that increased synergy will catalyze advancements in gene fusion identification, characterization and significance evaluation. Paper by Natasha Latysheva and M. Madan Babu can be found here.