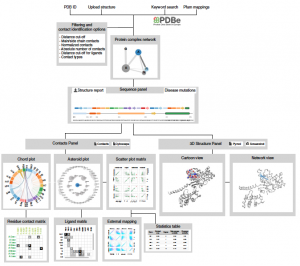
Visualizations of biomolecular structures empower us to gain insights into biological functions, generate testable hypotheses, and communicate biological concepts. Typical visualizations (such as ball and stick) primarily depict covalent bonds. In contrast, non-covalent contacts between atoms, which govern normal physiology, pathogenesis, and drug action, are seldom visualized. We present the Protein Contacts Atlas, an interactive resource of non-covalent contacts from over 100,000 PDB crystal structures. We developed multiple representations for visualization and analysis of non-covalent contacts at different scales of organization: atoms, residues, secondary structure, subunits, and entire complexes. The Protein Contacts Atlas enables researchers from different disciplines to investigate diverse questions in the framework of non-covalent contacts, including the interpretation of allostery, disease mutations and polymorphisms, by exploring individual subunits, interfaces, and protein–ligand contacts and by mapping external information. The Protein Contacts Atlas is available at http://www.mrc-lmb.cam.ac.uk/pca/ and also through PDBe.