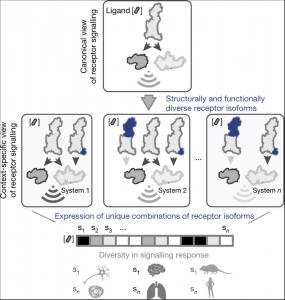
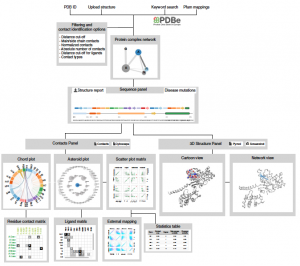
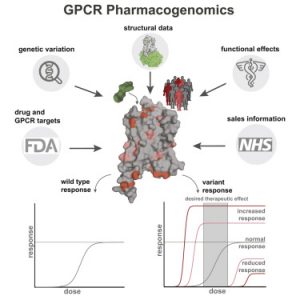
G-protein-coupled receptors (GPCRs) are membrane proteins that modulate physiology across human tissues in response to extracellular signals. GPCR-mediated signalling can differ because of changes in the sequence or expression of the receptors, leading to signalling bias when comparing diverse physiological systems. An underexplored source of such bias is the generation of functionally diverse GPCR isoforms with different patterns of expression across different tissues. Here we integrate data from human tissue-level transcriptomes, GPCR sequences and structures, proteomics, single-cell transcriptomics, population-wide genetic association studies and pharmacological experiments. We show how a single GPCR gene can diversify into several isoforms with distinct signalling properties, and how unique isoform combinations expressed in different tissues can generate distinct signalling states. Depending on their structural changes and expression patterns, some of the detected isoforms may influence cellular responses to drugs and represent new targets for developing drugs with improved tissue selectivity. Our findings highlight the need to move from a canonical to a context-specific view of GPCR signalling that considers how combinatorial expression of isoforms in a particular cell type, tissue or organism collectively influences receptor signalling and drug responses.
The paper by Maria Marti-Solano et al can be found here