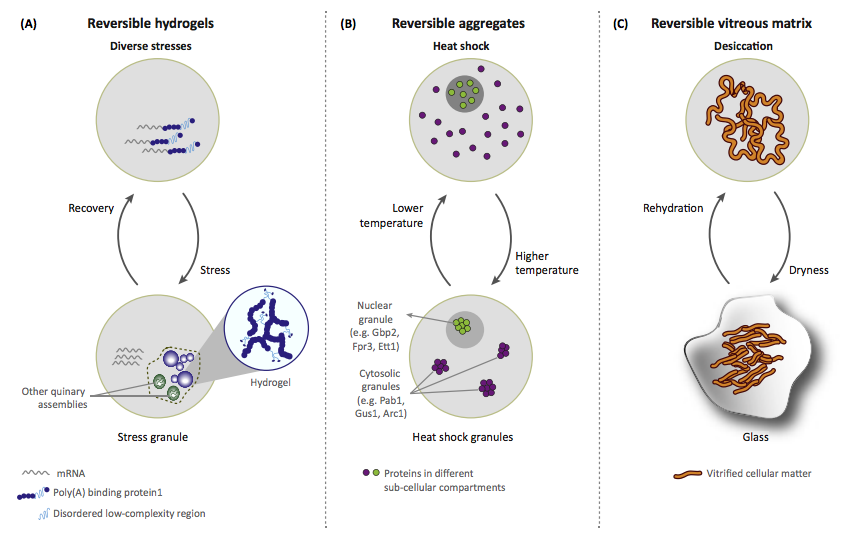
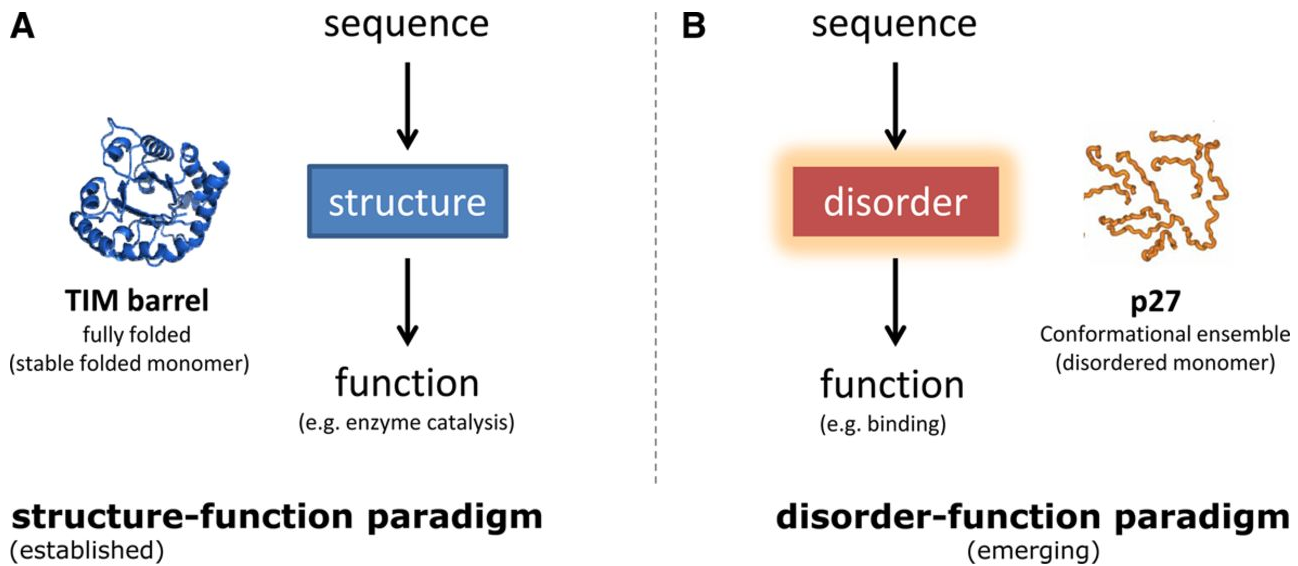
We will soon be recruiting at the PhD level in the following areas: Cancer Genetics, Human Genetics, Omics, Data Science, Synthetic Biology and Molecular/Cell Biology. Watch this space for future job postings, and follow Madan on Twitter.
Current job openings:
We are always looking for talented individuals who are motivated to explore the intersection between data science and biology in a multidisciplinary and diverse working environment. The Babu Group strives to build and maintain a culture of respect, diversity, and inclusion. We strongly feel these values directly contribute to the dynamic and intellectually stimulating scientific environment we can provide.