Dr. Balaji Santhanam from Harvard University has just started as a new Senior Investigator Scientist at the LMB. A very warm welcome from all of us Balaji. We are looking forward to some very exciting discussions and science in the coming days.

Dr. Balaji Santhanam from Harvard University has just started as a new Senior Investigator Scientist at the LMB. A very warm welcome from all of us Balaji. We are looking forward to some very exciting discussions and science in the coming days.

Elisabetta and James are our two new summer students who started this month. A very warm welcome to both of you and we look forward to some exciting science in the next couple of months!


Through a systematic effort that integrated multiple genome-scale datasets describing various aspects of DNA binding proteins, we show that activators tend to have binding site similarity to nucleosomal histones, resulting in competition for binding to DNA, while repressors show the opposite trend. You can read the paper here and the LMB press release here.
In this work, we show that nuclear rearrangement of repressive histone marks H3K9me3 and H3K27me3 into nonoverlapping structural layers characterizes senescence-associated heterochromatic foci (SAHF) formation in human fibroblasts. The experiments reveal that high-order heterochromatin formation and epigenetic remodeling of the genome can be discrete events. The paper by Chandra et al. can be read here.
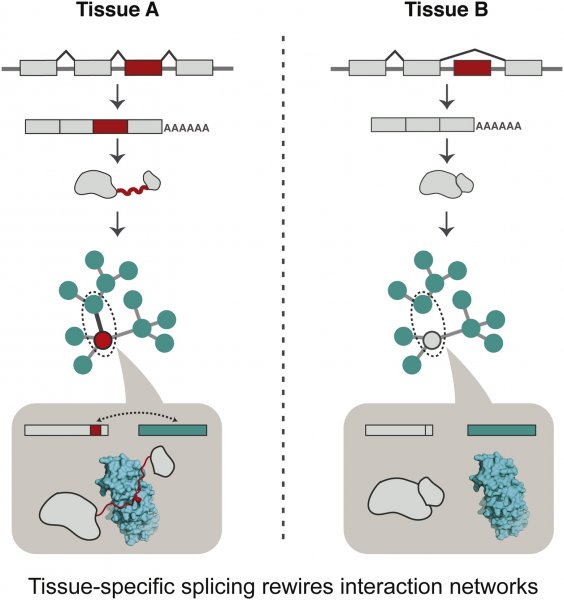
Our work on how tissue-specifically spliced segments can rewire protein interactions through disordered segments has been published in Molecular Cell. This work, which has been carried out over the last three years integrates multiple large-scale dataset to arrive at a general principle on how splicing can rewire interactions. The work received coverage in Nature Reviews Genetics and the Gates Trust. The press release on the LMB website can be read here.
Co-evolving positions within protein sequences have been used as spatial constraints to develop a computational approach for modeling membrane protein structures. In this mini-review, we discuss how co-evolving sites can be exploited for understanding protein structure and function. The paper can be read here.
The review on interplay between gene expression stochasticity and network was featured as a cover article in Trends in Genetics. In this work, we discuss and present new ideas on how gene expression noise can be tolerated, buffered or amplified by transcriptional networks. You can read the news item released by the Gates Trust here.