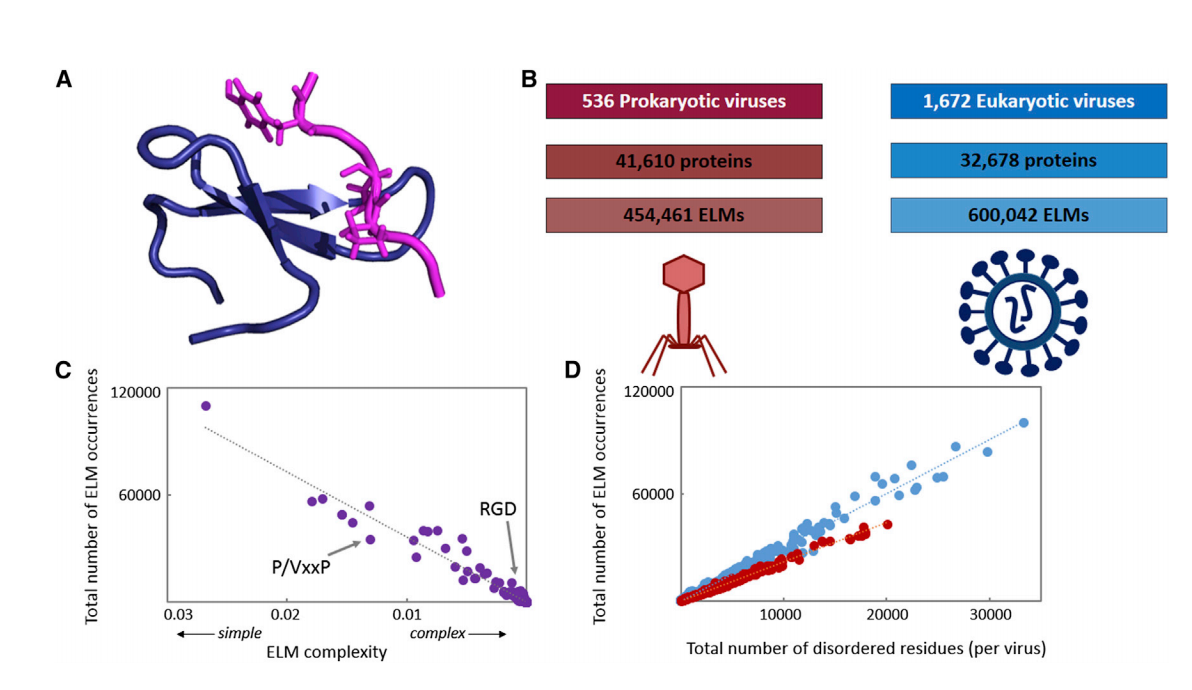
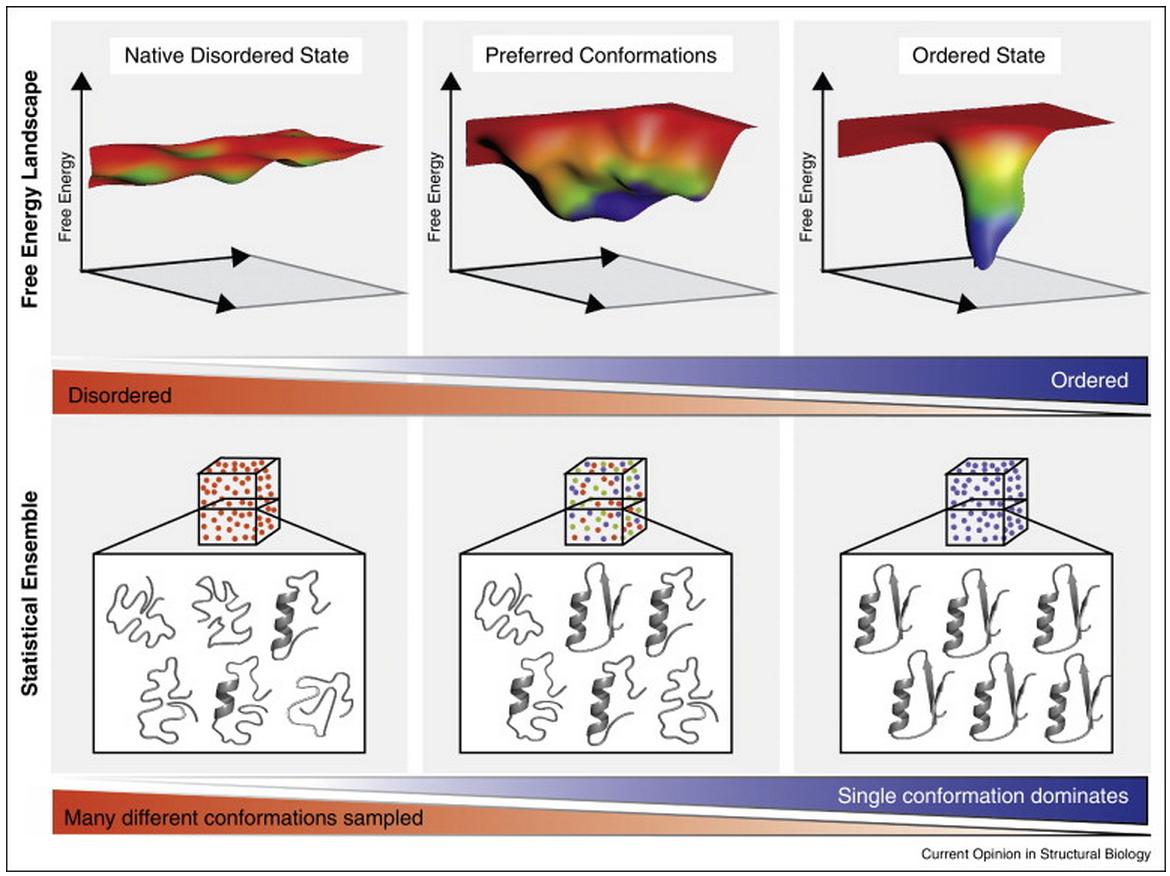
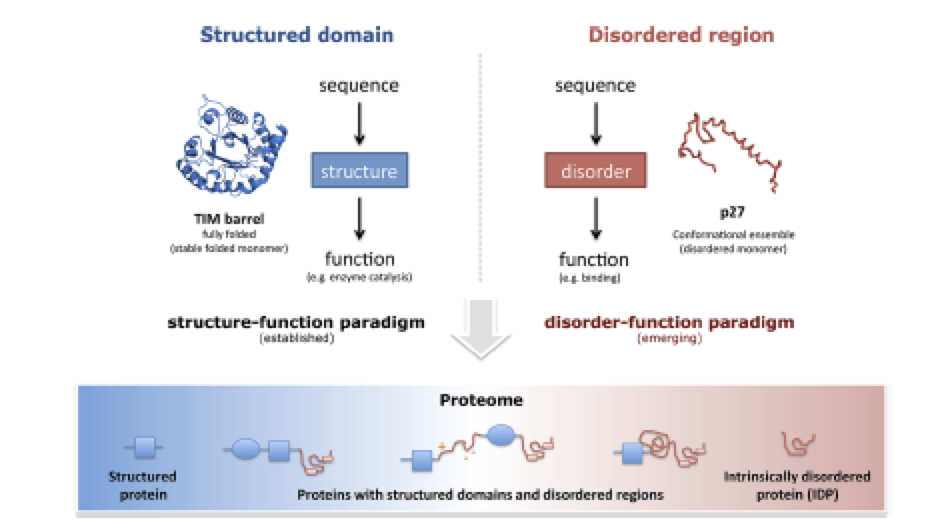
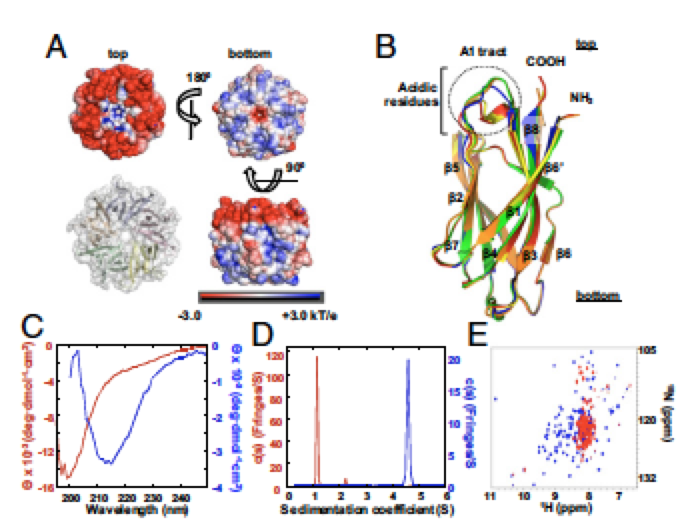
Intrinsically Disordered Segments Affect Protein Half-Life in the Cell and during Evolution
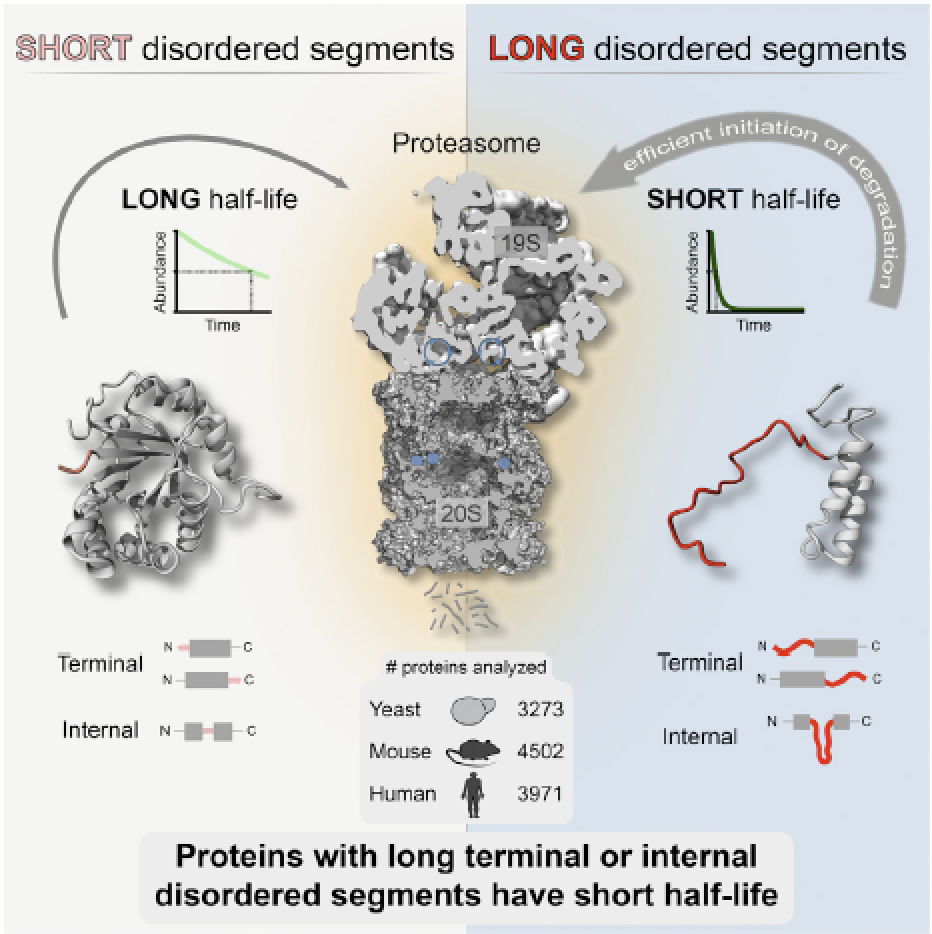
Precise control of protein turnover is essential for cellular homeostasis. The ubiquitin-proteasome system is well established as a major regulator of protein degradation, but an understanding of how inherent structural features influence the lifetimes of proteins is lacking. We report that yeast, mouse, and human proteins with terminal or internal intrinsically disordered segments have significantly shorter half-lives than proteins without these features. The lengths of the disordered segments that affect protein half-life are compatible with the structure of the proteasome. Divergence in terminal and internal disordered segments in yeast proteins originating from gene duplication leads to significantly altered half-life. Many paralogs that are affected by such changes participate in signaling, where altered protein half-life will directly impact cellular processes and function. Thus, natural variation in the length and position of disordered segments may affect protein half-life and could serve as an underappreciated source of genetic variation with important phenotypic consequences. The paper by Robin van der Lee et al can be found here.