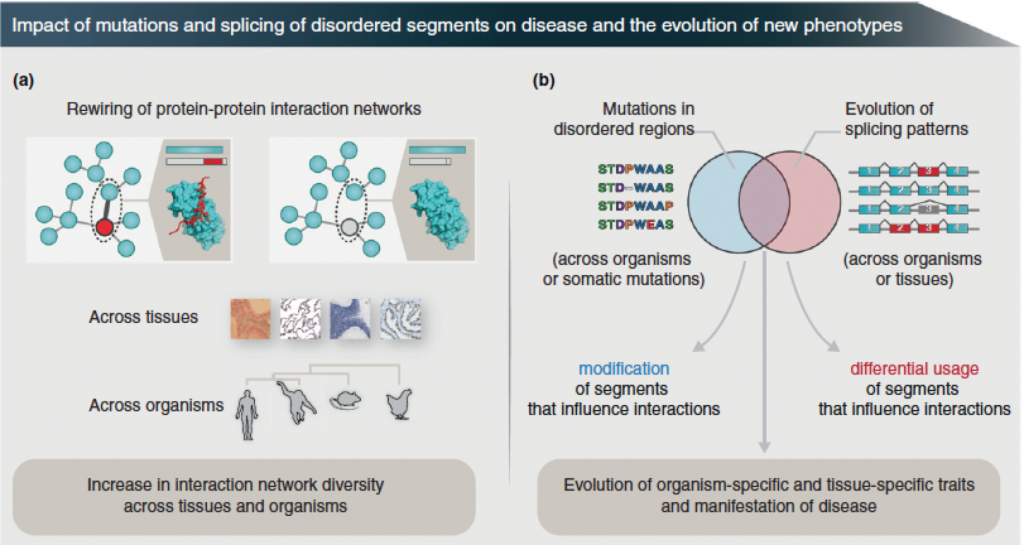
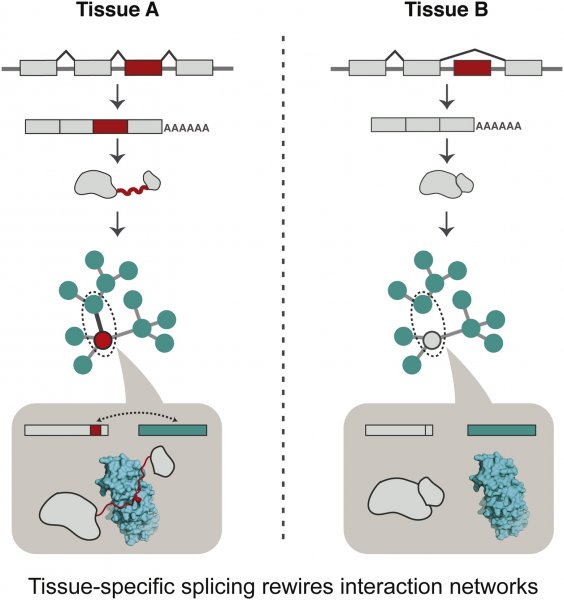
Alternative splicing of intrinsically disordered regions and rewiring of protein interactions
Alternatively spliced protein segments tend to be intrinsically disordered and contain linear interaction motifs and/or post-translational modification sites. An emerging concept is that differential inclusion of such disordered segments can mediate new protein interactions, and hence change the context in which the biochemical or molecular functions are carried out by the protein. Since genes with disordered regions are enriched in regulatory and signaling functions, the resulting protein isoforms could alter their function in different tissues and organisms by rewiring interaction networks through the recruitment of distinct interaction partners via the alternatively spliced disordered segments. In this manner, the alternative splicing of mRNA coding for disordered segments may contribute to the emergence of new traits during evolution, development and disease. The paper by Marija Buljan et al can be found here.