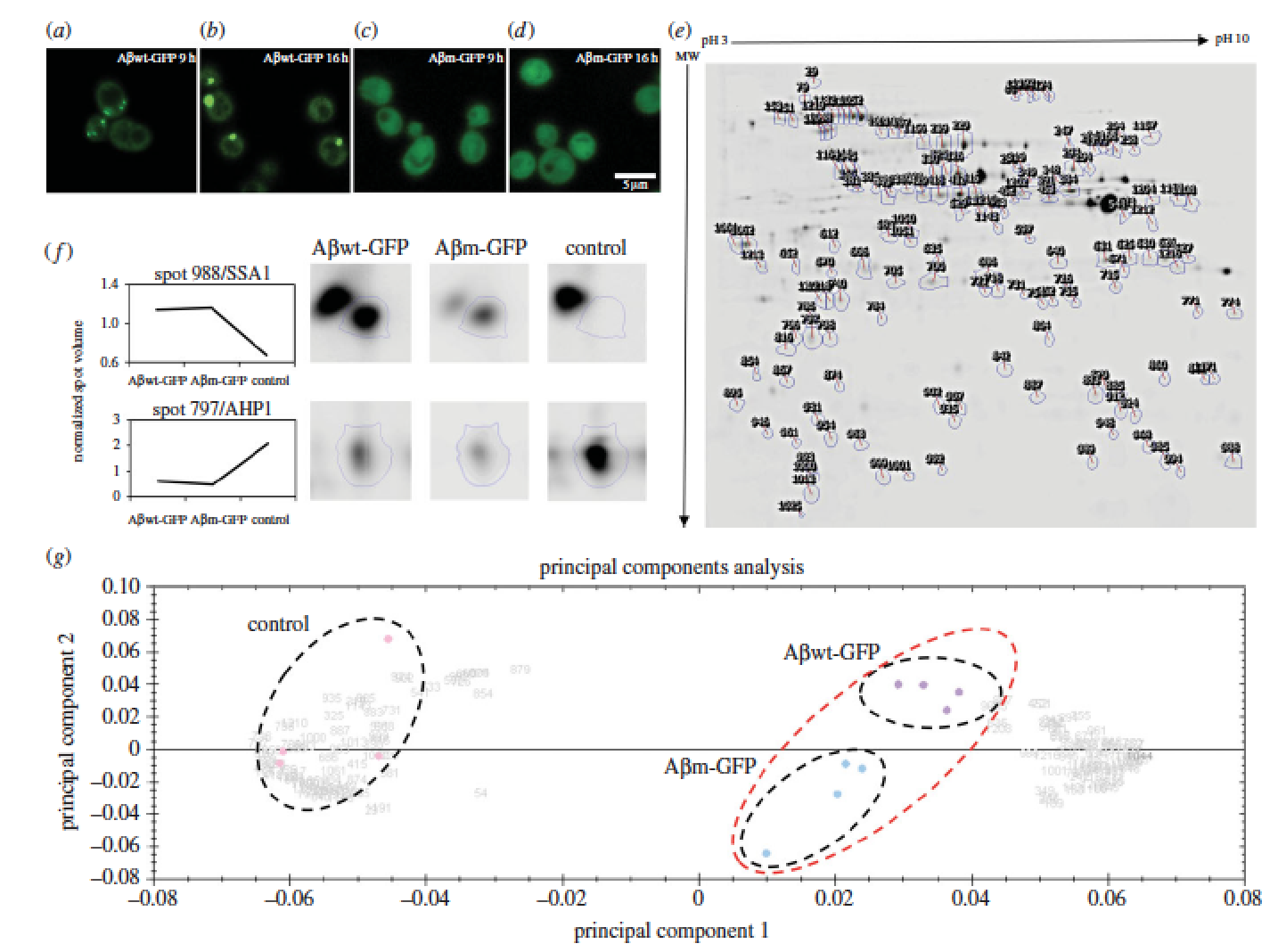
Proteins adopt defined structures and are crucial to most cellular functions. Their misfolding and aggregation is associated with numerous degenerative human disorders such as type II diabetes, Huntington’s or Alzheimer’s diseases. Here, we aim to understand why cells promote the formation of protein foci. Comparison of two amyloid-b-peptide variants, mostly insoluble but differently recruited by the cell (inclusion body versus diffused), reveals small differences in cell fitness and proteome response. We suggest that the levels of oxidative stress act as a sensor to trigger protein recruitment into foci. Our data support a common cytoplasmic response being able to discern and react to the specific properties of polypeptides. The paper by Natalia Sanchez de Groot can be viewed here.